Friday, 15 March, 2019
Tiny “blinkers” reveal molecules inside cells
Nanophysics

DNA-PAINT super-resolution image of all 124 DNA origami structures, color-coded according to the assigned binding frequency and spectral color. Credit: J.B. Woehrstein, F. Stehr, M.T. Strauss
LMU physicists led by Ralf Jungmann introduce an entirely new approach to super-resolution microscopy: Tiny ‘blinkers’ enable simultaneous imaging of multiple biomolecules.
In everyday life, blinking lights can send signals – for example, that a car is going to turn. Now, researchers have engineered tiny “blinkers” that reveal single molecules of RNA or protein inside cells based on the duration and frequency of each flash. The research, published in the journal Nano Letters, could allow scientists to see the locations of many different biomolecules in a cell simultaneously, possibly leading to better diagnostics and treatments.
Recently, scientists have developed super-resolution microscopes that can image single molecules that are only a few nanometers in size. To discriminate a specific nucleic acid or protein, they typically add a fluorescent probe that binds to that molecule and emits a certain wavelength of light. However, because the emission wavelengths of different fluorescent probes can overlap, researchers can usually only detect three or four unique proteins or nucleic acids at a time, instead of the thousands that exist in cells. Ralf Jungmann, Professor of Experimental Physics at LMU and head of the research group Molecular Imaging and Bionanotechnology at the Max Planck Institute of Biochemistry (Martinsried), and colleagues wondered if they could use fluorescent probes that blink with light at a variable duration and frequency to detect dozens of biomolecules at once. That way, they could use a single fluorophore to image many different molecules.
The researchers based their system on complementary sequences of DNA that come together to link a fluorophore with a target biomolecule and then fall apart again, generating a blinking fluorescent signal. By varying the length and number of DNA sequences bound to the target, the researchers could adjust how long the blink lasted, as well as how often blinks occurred. To test their approach in cells, the researchers imaged two different RNA molecules and two proteins. Then, they used three fluorescent probes to image 124 distinct DNA structures that contained different numbers of target DNAs so that they blinked at different frequencies. The procedure took only a few minutes and had an accuracy of 97.6 percent, the researchers say. (American Chemical Society/LMU)
Source: 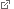 LMU press release
LMU press release LMU Pressemitteilung auf Deutsch
LMU Pressemitteilung auf Deutsch

